The genomic study of the horseshoe crab, which was done using NSCC’s supercomputing resources, can help in the conservation of the horseshoe crab and decipher its remarkably sensitive immune system.
The horseshoe crab has remained relatively unchanged after nearly half a billion years of evolutionary history. New genomic research led by scientists from A*STAR’s Institute of Molecular and Cell Biology (IMCB), with help from NUS and Republic Polytechnic researchers, have generated the most contiguous high-quality, chromosome-scale genome assembly for the mangrove horseshoe crab to date. The study was recently published in the journal Nature Communications.
The genomics study is a valuable resource for the conservation of this endangered species and is an important component in providing greater insight about the innate immune system of horseshoe crabs. A protein found in the blood of horseshoe crabs is used in the pharmaceutical and medical industry as an extremely sensitive agent for detecting endotoxin contamination.
“An important prerequisite for formulating effective conservation strategies is an understanding of genetic diversity and population structure but at present, there is no information about the population genetics of horseshoe crabs in Singapore,” said Professor Byrappa Venkatesh, who is the Research Director that heads the Comparative and Medical Genomics Laboratory at IMCB. Analysis of this highly-contiguous genome has revealed novel information about the evolution of the horseshoe crab including the possibility that there have been three rounds of whole-genome duplication in its lineage.
“Our study has shown that there was a marked decline in the population size of the mangrove horseshoe crab approximately 60,000 years ago coinciding with the onset of the recent ice age which has failed to recover due to the recent increased human activities such as the modification of coastal areas and overexploitation. Some suggestions for managing horseshoe crab stocks are habitat conservation, prohibiting horseshoe crab harvest, and protection of their breeding grounds,” added Professor Venkatesh.
Analysis of the mangrove horseshoe crab genome has identified many tandem gene clusters that are specific to its lineage including those that play important roles in the horseshoe crab’s innate immunity. For the horseshoe crabs to survive and propagate successfully in a mangrove environment where they are exposed to a variety of pathogens, they have evolved a sophisticated immune system response network for recognising, immobilising and killing of pathogens.
“The horseshoe crabs’ amebocytes are extremely sensitive to bacteria and hence is routinely used as a sensitive agent for detecting bacterial endotoxin,” explained Professor Venkatesh. “The expanded immune system gene families identified in our study are likely to be important components of the horseshoe crab’s immune response network which helps them mount a rapid and effective immune response to counteract pathogens.”
To achieve the chromosome-scale genome assembly study, Professor Venkatesh and his team leveraged NSCC’s high performance computing resources. “Our genome project used state-of-the-art ‘third-generation sequencing’ technology which involves single-molecule real time sequencing (SMRT) that generates reads of 20,000 base pairs (bp) and longer, which are necessary to provide the highest contiguity to the genome,” said Professor Venkatesh adding that the horseshoe crab genome is relatively large (approximately 2 Gb) and the team generated SMRT reads amounting to 100 times the size of the genome (200 Gb).
“The only way we could assemble the genome was by using the large number of high-performance nodes of the NSCC as conventional computational systems are incapable of assembling such a large amount of long reads. This allowed us to assemble the genome in a reasonable period and attempt several combinations of parameters to generate alternate assemblies that enabled us to obtain the best possible final genome assembly.”
For more information about the study, please refer to the paper entitled, “Chromosome-level assembly of the horseshoe crab genome provides insights into its genome evolution”.
NSCC NewsBytes June 2020
Other Case Studies
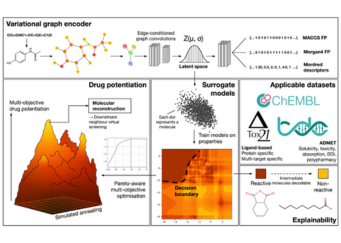
Advancing Drug Discovery Research using NSCC HPC resources
Researchers from Nanyang Technological University (NTU) are applying variational graph encoders as an effective generalist algorithm in computer-aided drug design (CADD)....
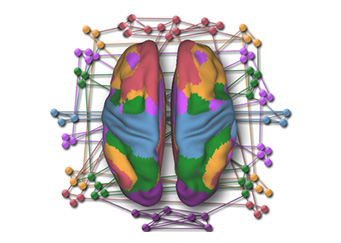
Gaining Deeper Insights into Mental Disorders through Brain Imaging and High-Performance Computing
Researchers from NUS are leveraging supercomputing to develop better strategies for prevention and treatment to mitigate the impact of mental illness. The human brain is a marvel...
Using Digital Twin Technology to Optimise the Industrial 3D Printing Process
Researchers from the Institute of High-Performance Computing (IHPC) are utilizing supercomputers to create a digital twin that furnishes users with comprehensive information...