To overcome the challenges of genomic data inconsistencies, the CSI Singapore team developed a Scalable Workflows for the Analysis of Genomes (SWAG), a high-performance bioinformatics pipeline deployed on NSCC Singapore’s ASPIRE 2A supercomputing system.
SWAG standardises the genomic data from diverse sources, ensuring that all datasets are processed with the same technical workflow. This eliminates errors and batch effects—variations introduced by different sequencing methods or tools—that can obscure the identification of true biological mutations. The harmonised data is then shared with National University of Singapore (NUS) research centres, supporting collaborative research in cancer genomics and facilitating the exchange of key insights and the development of innovative, data-driven treatments.
- Scalable Genomics Workflow (SWAG): A scalable HPC workflow that cleans and harmonises sequencing data from heterogeneous sources, eliminating batch effects and enabling reproducible scientific outcomes.
- High-Speed Data Transfer via SingAREN: Enabled fast downloads of petabytes of genomic data from the Genomic Data Commons (GDC) of the National Cancer Institute in the USA, significantly accelerating data accessibility and research timelines.
- NSCC Singapore’s ASPIRE 2A: Provided the massive compute power required to analyse complex datasets, apply AI/ML models, and simulate genome instability patterns using parallel processing capabilities.
The application of HPC in cancer genomics has had a profound impact on the research process. Key benefits include:
- Efficient Data Harmonisation: SWAG standardises datasets, ensuring that all genomic data is processed in a consistent and reproducible way. This eliminates inconsistencies, improving the reliability and accuracy of results.
- Faster Data Processing: The computational power of NSCC Singapore’s ASPIRE 2A enables the analysis of large genomic datasets in a fraction of the time it would take using traditional methods. This speeds up the overall research timeline, allowing for faster discoveries.
- AI-Driven Analysis: HPC facilitates the integration of AI models that can detect complex patterns in genomic data. These models help identify key genetic mutations that contribute to cancer, enabling more precise cancer predictions and the development of targeted treatments.
The success of this project has had far-reaching implications for cancer research in Singapore and beyond, including:
- Standardised Genomic Data: SWAG has set a new benchmark in Singapore and beyond for data processing in cancer genomics by eliminating batch effects, ensuring that data from different sources is harmonised for more reliable analysis.
- AI-Driven Cancer Predictions: The harmonised datasets are used to train discriminative AI, generative AI, and representational learning models, enabling more accurate identification of cancer-associated genetic changes. This paves the way for better diagnostics and targeted treatments.
- Collaborative Research: The clean datasets are shared across multiple research centres at NUS, fostering collaboration and accelerating progress in cancer research.
The development of SWAG, supported by NSCC Singapore’s HPC resources,have played a key role in significant breakthroughs in understanding the genetic causes of cancer. For instance, Prof. Ashok Venkitaraman’s lab, which collaborates closely with the Pitt lab, has made major advancements in understanding BRCA2, a key gene for breast and ovarian cancers. Mutations in BRCA2 and similar genes result in a distinct genetic signature, which can now be identified using AI models trained on genomic data. The capacity to analyse these vast datasets through the ASPIRE2A supercomputer has been instrumental in uncovering these genetic signatures. This breakthrough enables doctors to better predict which patients will benefit from targeted therapies, paving the way for more personalised cancer treatments.
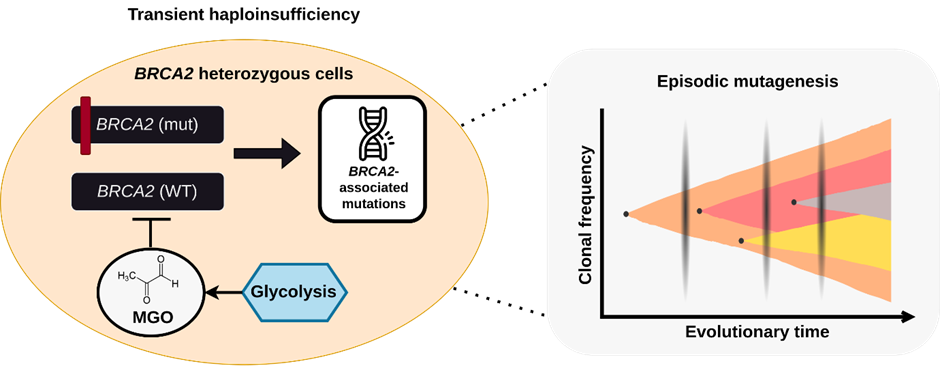
Caption: Graphical overview of metabolite-mediated BRCA2 haploinsufficiency and subsequent episodic mutagenesis within cells.
Additionally, the team also discovered that lifestyle factors such as poor diet, obesity, and diabetes can induce similar genetic mutations to those observed in BRCA2-related cancers. This finding suggests that lifestyle changes could reduce the risk of these mutations and lead to new prevention strategies. By understanding how these mutations occur, thanks to the computational power of HPC, researchers can develop better strategies for early cancer detection and treatment, ultimately improving patients’ chances for recovery.
“HPC has transformed how we process cancer genomic data. With NSCC Singapore’s ASPIRE 2A and SingAREN’s high-speed network, we can now analyse vast datasets more efficiently, eliminating batch effects and accelerating our ability to make meaningful discoveries. This research brings us one step closer to harnessing AI for cancer diagnostics and precision medicine.”

Dr. Jason J. Pitt
Principal Investigator, Head of Genomics and Data Analytics Core (GeDaC)
CSI Singapore





