Utilising high performance computing to understand the life cycle of Influenza A Virus to aid in the development and commercial production of vaccines.
Influenza A Virus (IAV) is a perennial public health threat that causes both seasonal epidemics and, less frequently, global pandemics. Statistics from the World Health Organisation (WHO) show that between three to five million severe IAV infections annually result in considerable economic losses, and more poignantly, nearly half a million deaths, particularly among the elderly, immunocompromised patients, and children.
The IAV genome consists of RNA and is divided into eight segments. All eight segments need to be packaged into a viral particle to form an infectious virus. However, the segmented genome allows for a process of re-assortment whereby segments are exchanged between infectious human strains and between human IAVs and animal IAVs, resulting in new virus strains of which there is no prior human immunity. Such strains may give rise to pandemic influenza.
A research team at A*STAR’s Bioinformatics Institute (BII) is making use of numerical simulations based on experimental data to understand the behaviour of the genomic segments and the role particular RNA structures play. The team is leveraging NSCC’s supercomputing resources to conduct coarse-grained and atomistic simulations of IAV segments that reveal the dynamics of these structures which will allow them to understand how these segments are interacting with one another during viral encapsidation.
This is important for the development and commercial production of vaccines where antigenic segments are combined with segments optimised for growth under laboratory conditions to maximise yield. Moreover, reassortment of viral genome segments is a key mechanism by which pandemic viruses cross species and hence understanding the molecular basis for this process will support efforts at early detection of dangerous mutations.
The team is now using the data derived from the simulations to parametrise ultra-coarse-grained models. This technique will allow them to observe genome assembly from the segments in real time and test computationally which parts of the genome drive virus packaging and re-assortment.
To find out more about the NSCC’s HPC resources and how you can tap on them, please contact [email protected].
NSCC NewsBytes April 2021
Other Case Studies
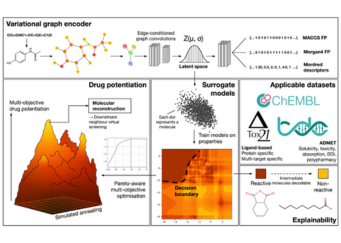
Advancing Drug Discovery Research using NSCC HPC resources
Researchers from Nanyang Technological University (NTU) are applying variational graph encoders as an effective generalist algorithm in computer-aided drug design (CADD)....
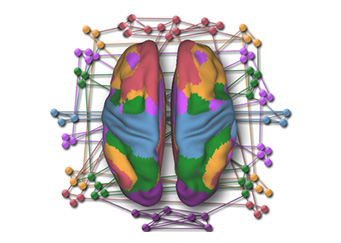
Gaining Deeper Insights into Mental Disorders through Brain Imaging and High-Performance Computing
Researchers from NUS are leveraging supercomputing to develop better strategies for prevention and treatment to mitigate the impact of mental illness. The human brain is a marvel...
Using Digital Twin Technology to Optimise the Industrial 3D Printing Process
Researchers from the Institute of High-Performance Computing (IHPC) are utilizing supercomputers to create a digital twin that furnishes users with comprehensive information...